Naive Bayes Classifier
May 13, 2019
This is the current version of the Naive Bayes workflow used to classify TA comments based on Subject and Structure. Features of Version 2r.2c:
- Data rows used for training and test datasets are assigned randomly but reproducibly using ‘set.seed’.
- Detailed final output matrix tables.
- Back-analysis evaluates both a testing sub-set and the entire input dataset.
- Outputs a tidy data table in CSV format with unmodified original comments, and hand-scored and NB-predicted Subject and Structure assignments.
Initial Setup
Calling required libraries.
library(tidyverse)
library(tm)
library(tidytext)
library(e1071)
library(caret)
library(ggplot2)Import the initial dataset. Then extract comments and classification data as subsets of CSV.
#Read in TA comments from CSV file.
base_dataVERB <- read_csv(file='data/coded_full_comments_dataset_Spring18anonVERB.csv')
#Select rows representing the sub-groups to compare.
comments_subsetVERB <- filter(base_dataVERB,code.subject=="1_basic"|code.subject=="2_writing"|code.subject=="3_technical"|code.subject=="4_logic")
#Reduce larger dataframe to 2 required columns of data, and put columns in order needed.
comments_rawVERB <- comments_subsetVERB %>% select(23,24,22)
#Rename the columns.
names(comments_rawVERB)[1] <- "subject"
names(comments_rawVERB)[2] <- "structure"
names(comments_rawVERB)[3] <- "text"
#Change "subject" element from character to a factor for analysis.
comments_rawVERB$subject <- factor(comments_rawVERB$subject)
str(comments_rawVERB$subject)## Factor w/ 4 levels "1_basic","2_writing",..: 3 2 3 3 3 3 4 4 4 3 ...table(comments_rawVERB$subject)##
## 1_basic 2_writing 3_technical 4_logic
## 211 2578 5409 1142#Change "structure" element from character to a factor for analysis.
comments_rawVERB$structure <- factor(comments_rawVERB$structure)
str(comments_rawVERB$structure)## Factor w/ 7 levels "1_pointer","2_copyedit",..: 2 4 4 3 2 2 5 4 5 3 ...table(comments_rawVERB$structure)##
## 1_pointer 2_copyedit 3_general 4_specific 6_holistic 7_idiomatic
## 91 2997 1881 3894 370 7
## 8_nobasis
## 100Randomize data used for training and testing sets. This block of code generates a reproducible vector of ~10,000 randomly assorted numbers using set.seed, then writes the vector back to data table.
#Adjust the number so it matches the number of rows in the original data frame
#Next, set the seed for random numbers, and randomize the order of the vector
vector <- 1:9340 #second value must match number of rows in working dataset
set.seed(123) #change this seed value, and the data are randomized in a new sequence.
vector <- sample(vector)
glimpse(vector)## int [1:9340] 2686 7362 3820 8245 8781 426 4930 8329 5146 4261 ...#Append the vector as a new column to "comments_rawVERB" with name "randomizer"
#Sort rows of dataframe according to "randomizer" column using dplyer 'arrange' function.
comments_rawVERB$randomizer <- c(vector)
comments_rawVERB_R <-comments_rawVERB %>% arrange(randomizer)
print(comments_rawVERB_R)## # A tibble: 9,340 x 4
## subject structure text randomizer
## <fct> <fct> <chr> <int>
## 1 3_technic… 4_specif… The commitment pulse occurs during the … 1
## 2 2_writing 2_copyed… This is not a scientific term 2
## 3 3_technic… 4_specif… Another potential reason could be error… 3
## 4 3_technic… 4_specif… I would rather focus on improving the p… 4
## 5 3_technic… 4_specif… What is the stimuli to open Ca++ channe… 5
## 6 3_technic… 2_copyed… Try something like, “The average R:S ra… 6
## 7 2_writing 4_specif… This goes in methods, but why’d you use… 7
## 8 2_writing 4_specif… Instead of saying that the obtained res… 8
## 9 3_technic… 2_copyed… Just refer to the study, not the author. 9
## 10 2_writing 1_pointer ? 10
## # … with 9,330 more rowsConvert data set to a volative corpus using “tm” library.
#Create the volatile corpus that contains the "text" vector from data frame.
comments_corpusVERB_R <- VCorpus(VectorSource(comments_rawVERB_R$text))
print(comments_corpusVERB_R)## <<VCorpus>>
## Metadata: corpus specific: 0, document level (indexed): 0
## Content: documents: 9340#Check out the first few text comments in the new corpus, which is basically a list that can be manipulated with list operations.
inspect(comments_corpusVERB_R[1:3])## <<VCorpus>>
## Metadata: corpus specific: 0, document level (indexed): 0
## Content: documents: 3
##
## [[1]]
## <<PlainTextDocument>>
## Metadata: 7
## Content: chars: 55
##
## [[2]]
## <<PlainTextDocument>>
## Metadata: 7
## Content: chars: 29
##
## [[3]]
## <<PlainTextDocument>>
## Metadata: 7
## Content: chars: 357#Use "as.character" function to see what a single text comment looks like.
as.character(comments_corpusVERB_R[[5]])## [1] "What is the stimuli to open Ca++ channels, the arrival of an AP or the opening of K+ channels? You should be precise."Text Data Transformations
The text data transforms in this code block are intentially separate “tm_map”" and “content_transformer” functions. This simplifies the process of testing how various combinations affect classification. Converting the text to lower case and removing punctuation are the baseline transformations. Switch between “{r}” and “{}” to turn other transformation blocks off or on as desired. Removing extra white spaces must be the last transformation, and is required.
#Convert text to all lower case letters.
comments_corpus_cleanVERB_R <- tm_map(comments_corpusVERB_R, content_transformer(tolower))
#Remove punctuation using the "removePunctuation" function.
#This step removes evidence of questions, so may remove data.
#THIS ALSO REMOVES EVIDENCE OF POINTER ITEMS
comments_corpus_cleanVERB_R <- tm_map(comments_corpus_cleanVERB_R, removePunctuation)
#Remove numerals.
#This step may remove evidence indicating technical comments
comments_corpus_cleanVERB_R <- tm_map(comments_corpus_cleanVERB_R, removeNumbers)
#Stopword removal.
#This is a standard cleanup step, but again may remove useful terms.
#The "stopwords" file is a convention; other files can be substituted for it
comments_corpus_cleanVERB_R <- tm_map(comments_corpus_cleanVERB_R, removeWords, stopwords())
#The option to apply stemming has been removed from this version. Stemming reduced accuracy in all trials. Future iterations should examine lemmatization instead.
#Final step removes white space from the document. This is NOT an optional step.
comments_corpus_cleanVERB_R <- tm_map(comments_corpus_cleanVERB_R, stripWhitespace)
#Look at an example of cleaned text comment to see if cleaned comment matches what is expected. Change the value inside double brackets to look at a different comment.
as.character((comments_corpus_cleanVERB_R[[5]]))## [1] " stimuli open ca channels arrival ap opening k channels precise"Tokenize the N-Grams
This code block uses tm’s NLP commands to assess different n-grams. For single words, change the “#” variable in “(words(x), #)” to “1”. Use other values to change the n-gram size. Data are stored in document-term matrix (dtm) format.
NLP_Tokenizer <- function(x) {
unlist(lapply(ngrams(words(x), 1), paste, collapse = " "), use.names = FALSE)
}
comments_dtm_1gram <- DocumentTermMatrix(comments_corpus_cleanVERB_R, control=list(tokenize = NLP_Tokenizer))
comments_dtm_1gram_tidy <- tidy(comments_dtm_1gram)
inspect(comments_dtm_1gram)## <<DocumentTermMatrix (documents: 9340, terms: 4837)>>
## Non-/sparse entries: 62771/45114809
## Sparsity : 100%
## Maximal term length: 69
## Weighting : term frequency (tf)
## Sample :
## Terms
## Docs can data don’t figure hypothesis include just need results use
## 1023 1 2 0 0 0 0 1 0 0 0
## 1700 1 0 0 0 1 1 1 0 0 0
## 1986 0 1 0 1 0 0 0 0 0 0
## 5606 0 0 0 0 1 0 0 0 0 0
## 582 3 0 0 0 0 0 0 0 0 0
## 5937 0 0 0 1 0 3 1 0 2 0
## 7257 1 1 0 2 0 0 0 0 1 0
## 7325 1 2 1 4 0 0 0 1 0 1
## 8865 1 1 1 0 0 1 0 1 0 0
## 9224 0 1 1 0 0 1 0 1 0 0Split Data to Training, Testing Sets
This code splits data into 75% training and 25% testing sets, so that after Naive Bayes algorithm is trained it can be tested on unseen data.
#This block is for convenience. It is not required if the number of training, test rows is known.
.75 * 9340 #Number of rows in the dataset; product is #rows for training
.25 * 9340 #Product is #rows for testing setThis code divides the previously randomized comments into training, testing, and re-analysis subsets of data.
comments_dtm_all <- comments_dtm_1gram[1:9340, ]
comments_dtm_train <- comments_dtm_1gram[1:7005, ]
comments_dtm_test <- comments_dtm_1gram[7006:9340, ]This saves 3 vectors containing labels for the rows in the training and testing vectors
comments_all_labels_subject <- comments_rawVERB_R[1:9340,]$subject
comments_train_labels_subject <- comments_rawVERB_R[1:7005, ]$subject
comments_test_labels_subject <- comments_rawVERB_R[7006:9340,]$subject
comments_all_labels_structure <- comments_rawVERB_R[1:9340,]$structure
comments_train_labels_structure <- comments_rawVERB_R[1:7005, ]$structure
comments_test_labels_structure <- comments_rawVERB_R[7006:9340,]$structureThis checks the proportions of sub-categories in training and testing groups. The proportion of each sub-category should be similar (<1% difference) between the full dataset, and training and testing data subsets.
prop.table(table(comments_all_labels_subject))## comments_all_labels_subject
## 1_basic 2_writing 3_technical 4_logic
## 0.02259101 0.27601713 0.57912206 0.12226981prop.table(table(comments_train_labels_subject))## comments_train_labels_subject
## 1_basic 2_writing 3_technical 4_logic
## 0.02226981 0.27551749 0.58129907 0.12091363prop.table(table(comments_test_labels_subject))## comments_test_labels_subject
## 1_basic 2_writing 3_technical 4_logic
## 0.0235546 0.2775161 0.5725910 0.1263383prop.table(table(comments_all_labels_structure))## comments_all_labels_structure
## 1_pointer 2_copyedit 3_general 4_specific 6_holistic
## 0.0097430407 0.3208779443 0.2013918630 0.4169164882 0.0396145610
## 7_idiomatic 8_nobasis
## 0.0007494647 0.0107066381prop.table(table(comments_train_labels_structure))## comments_train_labels_structure
## 1_pointer 2_copyedit 3_general 4_specific 6_holistic 7_idiomatic
## 0.009136331 0.325053533 0.200000000 0.415132049 0.039971449 0.000856531
## 8_nobasis
## 0.009850107prop.table(table(comments_test_labels_structure))## comments_test_labels_structure
## 1_pointer 2_copyedit 3_general 4_specific 6_holistic
## 0.0115631692 0.3083511777 0.2055674518 0.4222698073 0.0385438972
## 7_idiomatic 8_nobasis
## 0.0004282655 0.0132762313Preparing Data for Naive Bayes Training
Remove words from the document-term matrix that appear less than 5 times.
comments_freq_words <- findFreqTerms(comments_dtm_train, 5)
str(comments_freq_words)## chr [1:1330] "’ll" "’re" "’ve" "“correct”" "“figure" "“group" ...Limit document-term matrix to words in the comments_freq_words vector. We are using all of the rows, but we want to limit the columns to these words in the frequency vector.
comments_dtm_freq_all <- comments_dtm_all[ , comments_freq_words]
comments_dtm_freq_train <- comments_dtm_train[ , comments_freq_words]
comments_dtm_freq_test <- comments_dtm_test[ , comments_freq_words]The e1071 Naive Bayes classifier works with categorical features, so the matrix must be converted “yes” and “no” categorical variables. This is done using a convert_counts function and applying it to the data. This replaces values greater than 0 with yes, and values not greater than 0 with no.
convert_counts2 <- function(x) {
x <- ifelse(x > 0, "Yes", "No")
}The resulting matrices have cells indicating “yes” or “no” if the word represented by the column appears in the text comment represented by the row.
The block below is an alternate version of the convert_function code block that KEEPs word frequencies while still using the same variable names and code structure. The processed matrices have cells with word frequencies instead of categorical yes/no values. This block only works when performing binary classification (ingroup/outgroup). The classifier fails when frequency values are used with data in multiple categories.
convert_counts2 <- function(x) {
y <- x
}This block applies whichever “convert_counts2” function is currently active.
comments_all <- apply(comments_dtm_freq_all, MARGIN = 2, convert_counts2)
comments_train <- apply(comments_dtm_freq_train, MARGIN = 2, convert_counts2)
comments_test <- apply(comments_dtm_freq_test, MARGIN = 2, convert_counts2)Train Model, Predict & Evaluate for Subject
This block predicts whether a message is likely to be in group 1_basic, 2_writing, 3_technical, or 4_logic.
# Train the classifier on training data, then test on test dataset, and full dataset.
comments_classifier_subject <- naiveBayes(comments_train, comments_train_labels_subject, laplace=1)
comments_test_pred_subject <- predict(comments_classifier_subject, comments_test)
comments_all_pred_subject <- predict(comments_classifier_subject, comments_all)comments_classifier_structure <- naiveBayes(comments_train, comments_train_labels_structure, laplace=1)
comments_test_pred_structure <- predict(comments_classifier_structure, comments_test)
comments_all_pred_structure <- predict(comments_classifier_structure, comments_all)A truth table lists how many of the predicted categories are in their correct categories. Adding diagonally and dividing by total count gives the percent accuracy at predicting Subject.
# Create a truth table by tabulating the predicted class labels with the actual class labels
table("Predictions"= comments_test_pred_subject, "Actual Subject Test" = comments_test_labels_subject )## Actual Subject Test
## Predictions 1_basic 2_writing 3_technical 4_logic
## 1_basic 28 3 11 0
## 2_writing 11 479 113 70
## 3_technical 14 119 1168 58
## 4_logic 2 47 45 167table("Predictions"= comments_all_pred_subject, "Actual Subject All" = comments_all_labels_subject )## Actual Subject All
## Predictions 1_basic 2_writing 3_technical 4_logic
## 1_basic 100 10 38 4
## 2_writing 54 1952 369 246
## 3_technical 49 460 4819 179
## 4_logic 8 156 183 713table("Predictions"= comments_test_pred_structure, "Actual Structure Test" = comments_test_labels_structure )## Actual Structure Test
## Predictions 1_pointer 2_copyedit 3_general 4_specific 6_holistic
## 1_pointer 0 2 0 3 0
## 2_copyedit 27 583 129 271 19
## 3_general 0 43 277 121 11
## 4_specific 0 91 66 538 37
## 6_holistic 0 1 8 51 23
## 7_idiomatic 0 0 0 0 0
## 8_nobasis 0 0 0 2 0
## Actual Structure Test
## Predictions 7_idiomatic 8_nobasis
## 1_pointer 0 0
## 2_copyedit 0 25
## 3_general 1 6
## 4_specific 0 0
## 6_holistic 0 0
## 7_idiomatic 0 0
## 8_nobasis 0 0table("Predictions"= comments_all_pred_structure, "Actual Structure All" = comments_all_labels_structure )## Actual Structure All
## Predictions 1_pointer 2_copyedit 3_general 4_specific 6_holistic
## 1_pointer 0 6 0 12 0
## 2_copyedit 90 2522 440 1023 52
## 3_general 0 133 1167 414 48
## 4_specific 1 320 245 2277 114
## 6_holistic 0 16 28 156 156
## 7_idiomatic 0 0 0 0 0
## 8_nobasis 0 0 1 12 0
## Actual Structure All
## Predictions 7_idiomatic 8_nobasis
## 1_pointer 0 0
## 2_copyedit 2 86
## 3_general 1 12
## 4_specific 4 2
## 6_holistic 0 0
## 7_idiomatic 0 0
## 8_nobasis 0 0#An alternative is to use the Cross Table command (from "gmodels" package) to generate a more detailed output table with frequency values for each individual cell.
#CrossTable(comments_test_pred_subject, comments_test_labels_subject, prop.chisq = FALSE, prop.t = FALSE, dnn = c('predicted', 'actual'))The faster method is to calculate a confusion matrix, which displays BOTH the truth table, and the final overall accuracy. A confusion matrix can be queried for specific values.
# Prepare the confusion matrix.
conf.mat1 <- confusionMatrix(comments_test_pred_subject, comments_test_labels_subject)
conf.mat2 <- confusionMatrix(comments_all_pred_subject, comments_all_labels_subject)
conf.mat3 <- confusionMatrix(comments_test_pred_structure, comments_test_labels_structure)
conf.mat4 <- confusionMatrix(comments_all_pred_structure, comments_all_labels_structure)
#These commands show how to print out individual pieces of the confusion matrix. They show both the overall and by-group accuracy values for Subject x Test Subset.
print ("Test, Subject")## [1] "Test, Subject"conf.mat1## Confusion Matrix and Statistics
##
## Reference
## Prediction 1_basic 2_writing 3_technical 4_logic
## 1_basic 28 3 11 0
## 2_writing 11 479 113 70
## 3_technical 14 119 1168 58
## 4_logic 2 47 45 167
##
## Overall Statistics
##
## Accuracy : 0.7889
## 95% CI : (0.7717, 0.8053)
## No Information Rate : 0.5726
## P-Value [Acc > NIR] : < 2e-16
##
## Kappa : 0.631
## Mcnemar's Test P-Value : 0.03925
##
## Statistics by Class:
##
## Class: 1_basic Class: 2_writing Class: 3_technical
## Sensitivity 0.50909 0.7392 0.8736
## Specificity 0.99386 0.8850 0.8086
## Pos Pred Value 0.66667 0.7117 0.8595
## Neg Pred Value 0.98823 0.8983 0.8268
## Prevalence 0.02355 0.2775 0.5726
## Detection Rate 0.01199 0.2051 0.5002
## Detection Prevalence 0.01799 0.2882 0.5820
## Balanced Accuracy 0.75148 0.8121 0.8411
## Class: 4_logic
## Sensitivity 0.56610
## Specificity 0.95392
## Pos Pred Value 0.63985
## Neg Pred Value 0.93828
## Prevalence 0.12634
## Detection Rate 0.07152
## Detection Prevalence 0.11178
## Balanced Accuracy 0.76001# This block outputs the by-group accuracy values in a single column for Subject x Test Subset.
print ("Test, Subject")## [1] "Test, Subject"conf.mat1$byClass## Sensitivity Specificity Pos Pred Value Neg Pred Value
## Class: 1_basic 0.5090909 0.9938596 0.6666667 0.9882250
## Class: 2_writing 0.7391975 0.8850030 0.7117385 0.8983153
## Class: 3_technical 0.8735976 0.8086172 0.8594555 0.8268443
## Class: 4_logic 0.5661017 0.9539216 0.6398467 0.9382835
## Precision Recall F1 Prevalence Detection Rate
## Class: 1_basic 0.6666667 0.5090909 0.5773196 0.0235546 0.01199143
## Class: 2_writing 0.7117385 0.7391975 0.7252082 0.2775161 0.20513919
## Class: 3_technical 0.8594555 0.8735976 0.8664688 0.5725910 0.50021413
## Class: 4_logic 0.6398467 0.5661017 0.6007194 0.1263383 0.07152034
## Detection Prevalence Balanced Accuracy
## Class: 1_basic 0.01798715 0.7514753
## Class: 2_writing 0.28822270 0.8121002
## Class: 3_technical 0.58201285 0.8411074
## Class: 4_logic 0.11177730 0.7600116# This block outputs a single line table with overall accuracy, kappa, lower and higher limits of accuracy, accuracy null value, P value for accuracy, and Mcnemar P value for Subject x Test Subset.
print ("Test, Subject")## [1] "Test, Subject"conf.mat1$overall## Accuracy Kappa AccuracyLower AccuracyUpper AccuracyNull
## 7.888651e-01 6.310203e-01 7.717402e-01 8.052621e-01 5.725910e-01
## AccuracyPValue McnemarPValue
## 1.467772e-107 3.925158e-02# This block outputs JUST the overall accuracy value for Subject x Test Subset.
print ("Test, Subject")## [1] "Test, Subject"conf.mat1$overall['Accuracy']## Accuracy
## 0.7888651# This block outputs both the overall and by-group accuracy values for Subject x All Data.
print ("All, Subject")## [1] "All, Subject"conf.mat2## Confusion Matrix and Statistics
##
## Reference
## Prediction 1_basic 2_writing 3_technical 4_logic
## 1_basic 100 10 38 4
## 2_writing 54 1952 369 246
## 3_technical 49 460 4819 179
## 4_logic 8 156 183 713
##
## Overall Statistics
##
## Accuracy : 0.812
## 95% CI : (0.8039, 0.8199)
## No Information Rate : 0.5791
## P-Value [Acc > NIR] : < 2.2e-16
##
## Kappa : 0.6683
## Mcnemar's Test P-Value : 1.025e-11
##
## Statistics by Class:
##
## Class: 1_basic Class: 2_writing Class: 3_technical
## Sensitivity 0.47393 0.7572 0.8909
## Specificity 0.99430 0.9011 0.8250
## Pos Pred Value 0.65789 0.7448 0.8751
## Neg Pred Value 0.98792 0.9068 0.8461
## Prevalence 0.02259 0.2760 0.5791
## Detection Rate 0.01071 0.2090 0.5160
## Detection Prevalence 0.01627 0.2806 0.5896
## Balanced Accuracy 0.73412 0.8291 0.8580
## Class: 4_logic
## Sensitivity 0.62434
## Specificity 0.95767
## Pos Pred Value 0.67264
## Neg Pred Value 0.94819
## Prevalence 0.12227
## Detection Rate 0.07634
## Detection Prevalence 0.11349
## Balanced Accuracy 0.79101# This block outputs by-group accuracy values in a single column for Subject x All Data.
print ("All, Subject")## [1] "All, Subject"conf.mat2$byClass## Sensitivity Specificity Pos Pred Value Neg Pred Value
## Class: 1_basic 0.4739336 0.9943039 0.6578947 0.9879190
## Class: 2_writing 0.7571761 0.9010648 0.7447539 0.9068314
## Class: 3_technical 0.8909225 0.8249809 0.8750681 0.8460736
## Class: 4_logic 0.6243433 0.9576726 0.6726415 0.9481884
## Precision Recall F1 Prevalence Detection Rate
## Class: 1_basic 0.6578947 0.4739336 0.5509642 0.02259101 0.01070664
## Class: 2_writing 0.7447539 0.7571761 0.7509136 0.27601713 0.20899358
## Class: 3_technical 0.8750681 0.8909225 0.8829241 0.57912206 0.51595289
## Class: 4_logic 0.6726415 0.6243433 0.6475931 0.12226981 0.07633833
## Detection Prevalence Balanced Accuracy
## Class: 1_basic 0.01627409 0.7341188
## Class: 2_writing 0.28062099 0.8291204
## Class: 3_technical 0.58961456 0.8579517
## Class: 4_logic 0.11349036 0.7910079# This block outputs a single line table with overall accuracy, kappa, lower and higher limits of accuracy, accuracy null value, P value for accuracy, and Mcnemar P value for Subject x All Data.
print ("All, Subject")## [1] "All, Subject"conf.mat2$overall## Accuracy Kappa AccuracyLower AccuracyUpper AccuracyNull
## 8.119914e-01 6.683222e-01 8.039172e-01 8.198706e-01 5.791221e-01
## AccuracyPValue McnemarPValue
## 0.000000e+00 1.025480e-11# This block outputs JUST the overall accuracy value for Subject x All Data.
print ("All, Subject")## [1] "All, Subject"conf.mat2$overall['Accuracy']## Accuracy
## 0.8119914# This block outputs both the overall and by-group accuracy values for Structure x Test Subset.
print ("Test, Structure")## [1] "Test, Structure"conf.mat3## Confusion Matrix and Statistics
##
## Reference
## Prediction 1_pointer 2_copyedit 3_general 4_specific 6_holistic
## 1_pointer 0 2 0 3 0
## 2_copyedit 27 583 129 271 19
## 3_general 0 43 277 121 11
## 4_specific 0 91 66 538 37
## 6_holistic 0 1 8 51 23
## 7_idiomatic 0 0 0 0 0
## 8_nobasis 0 0 0 2 0
## Reference
## Prediction 7_idiomatic 8_nobasis
## 1_pointer 0 0
## 2_copyedit 0 25
## 3_general 1 6
## 4_specific 0 0
## 6_holistic 0 0
## 7_idiomatic 0 0
## 8_nobasis 0 0
##
## Overall Statistics
##
## Accuracy : 0.6086
## 95% CI : (0.5884, 0.6284)
## No Information Rate : 0.4223
## P-Value [Acc > NIR] : < 2.2e-16
##
## Kappa : 0.4299
## Mcnemar's Test P-Value : NA
##
## Statistics by Class:
##
## Class: 1_pointer Class: 2_copyedit Class: 3_general
## Sensitivity 0.000000 0.8097 0.5771
## Specificity 0.997834 0.7084 0.9019
## Pos Pred Value 0.000000 0.5531 0.6035
## Neg Pred Value 0.988412 0.8931 0.8918
## Prevalence 0.011563 0.3084 0.2056
## Detection Rate 0.000000 0.2497 0.1186
## Detection Prevalence 0.002141 0.4514 0.1966
## Balanced Accuracy 0.498917 0.7590 0.7395
## Class: 4_specific Class: 6_holistic
## Sensitivity 0.5456 0.25556
## Specificity 0.8562 0.97327
## Pos Pred Value 0.7350 0.27711
## Neg Pred Value 0.7205 0.97025
## Prevalence 0.4223 0.03854
## Detection Rate 0.2304 0.00985
## Detection Prevalence 0.3135 0.03555
## Balanced Accuracy 0.7009 0.61441
## Class: 7_idiomatic Class: 8_nobasis
## Sensitivity 0.0000000 0.0000000
## Specificity 1.0000000 0.9991319
## Pos Pred Value NaN 0.0000000
## Neg Pred Value 0.9995717 0.9867124
## Prevalence 0.0004283 0.0132762
## Detection Rate 0.0000000 0.0000000
## Detection Prevalence 0.0000000 0.0008565
## Balanced Accuracy 0.5000000 0.4995660# This block outputs by-group accuracy values in a single column for Structure x Test Subset.
print ("Test, Structure")## [1] "Test, Structure"conf.mat3$byClass## Sensitivity Specificity Pos Pred Value Neg Pred Value
## Class: 1_pointer 0.0000000 0.9978336 0.0000000 0.9884120
## Class: 2_copyedit 0.8097222 0.7083591 0.5531309 0.8930523
## Class: 3_general 0.5770833 0.9018868 0.6034858 0.8917910
## Class: 4_specific 0.5456389 0.8561898 0.7349727 0.7205240
## Class: 6_holistic 0.2555556 0.9732739 0.2771084 0.9702487
## Class: 7_idiomatic 0.0000000 1.0000000 NaN 0.9995717
## Class: 8_nobasis 0.0000000 0.9991319 0.0000000 0.9867124
## Precision Recall F1 Prevalence
## Class: 1_pointer 0.0000000 0.0000000 NaN 0.0115631692
## Class: 2_copyedit 0.5531309 0.8097222 0.6572717 0.3083511777
## Class: 3_general 0.6034858 0.5770833 0.5899894 0.2055674518
## Class: 4_specific 0.7349727 0.5456389 0.6263097 0.4222698073
## Class: 6_holistic 0.2771084 0.2555556 0.2658960 0.0385438972
## Class: 7_idiomatic NA 0.0000000 NA 0.0004282655
## Class: 8_nobasis 0.0000000 0.0000000 NaN 0.0132762313
## Detection Rate Detection Prevalence Balanced Accuracy
## Class: 1_pointer 0.000000000 0.002141328 0.4989168
## Class: 2_copyedit 0.249678801 0.451391863 0.7590407
## Class: 3_general 0.118629550 0.196573876 0.7394851
## Class: 4_specific 0.230406852 0.313490364 0.7009144
## Class: 6_holistic 0.009850107 0.035546039 0.6144147
## Class: 7_idiomatic 0.000000000 0.000000000 0.5000000
## Class: 8_nobasis 0.000000000 0.000856531 0.4995660# This block outputs a single line table with overall accuracy, kappa, lower and higher limits of accuracy, accuracy null value, P value for accuracy, and Mcnemar P value for Structure x Test Subset.
print ("Test, Structure")## [1] "Test, Structure"conf.mat3$overall## Accuracy Kappa AccuracyLower AccuracyUpper AccuracyNull
## 6.085653e-01 4.299106e-01 5.884285e-01 6.284293e-01 4.222698e-01
## AccuracyPValue McnemarPValue
## 3.105959e-73 NaN# This block outputs JUST the overall accuracy value for Structure x Test Subset.
conf.mat3$overall['Accuracy']## Accuracy
## 0.6085653print ("Test, Structure")## [1] "Test, Structure"# This block outputs both the overall and by-group accuracy values for Structure x All Data.
print ("All, Structure")## [1] "All, Structure"conf.mat4## Confusion Matrix and Statistics
##
## Reference
## Prediction 1_pointer 2_copyedit 3_general 4_specific 6_holistic
## 1_pointer 0 6 0 12 0
## 2_copyedit 90 2522 440 1023 52
## 3_general 0 133 1167 414 48
## 4_specific 1 320 245 2277 114
## 6_holistic 0 16 28 156 156
## 7_idiomatic 0 0 0 0 0
## 8_nobasis 0 0 1 12 0
## Reference
## Prediction 7_idiomatic 8_nobasis
## 1_pointer 0 0
## 2_copyedit 2 86
## 3_general 1 12
## 4_specific 4 2
## 6_holistic 0 0
## 7_idiomatic 0 0
## 8_nobasis 0 0
##
## Overall Statistics
##
## Accuracy : 0.6555
## 95% CI : (0.6457, 0.6651)
## No Information Rate : 0.4169
## P-Value [Acc > NIR] : < 2.2e-16
##
## Kappa : 0.4956
## Mcnemar's Test P-Value : NA
##
## Statistics by Class:
##
## Class: 1_pointer Class: 2_copyedit Class: 3_general
## Sensitivity 0.000000 0.8415 0.6204
## Specificity 0.998054 0.7331 0.9185
## Pos Pred Value 0.000000 0.5983 0.6575
## Neg Pred Value 0.990238 0.9073 0.9056
## Prevalence 0.009743 0.3209 0.2014
## Detection Rate 0.000000 0.2700 0.1249
## Detection Prevalence 0.001927 0.4513 0.1900
## Balanced Accuracy 0.499027 0.7873 0.7695
## Class: 4_specific Class: 6_holistic
## Sensitivity 0.5847 0.42162
## Specificity 0.8740 0.97770
## Pos Pred Value 0.7685 0.43820
## Neg Pred Value 0.7464 0.97618
## Prevalence 0.4169 0.03961
## Detection Rate 0.2438 0.01670
## Detection Prevalence 0.3172 0.03812
## Balanced Accuracy 0.7294 0.69966
## Class: 7_idiomatic Class: 8_nobasis
## Sensitivity 0.0000000 0.000000
## Specificity 1.0000000 0.998593
## Pos Pred Value NaN 0.000000
## Neg Pred Value 0.9992505 0.989278
## Prevalence 0.0007495 0.010707
## Detection Rate 0.0000000 0.000000
## Detection Prevalence 0.0000000 0.001392
## Balanced Accuracy 0.5000000 0.499297# This block outputs by-group accuracy values in a single column for Structure x All Data.
print ("All, Structure")## [1] "All, Structure"conf.mat4$byClass## Sensitivity Specificity Pos Pred Value Neg Pred Value
## Class: 1_pointer 0.0000000 0.9980538 0.0000000 0.9902381
## Class: 2_copyedit 0.8415082 0.7330916 0.5983393 0.9073171
## Class: 3_general 0.6204147 0.9184877 0.6574648 0.9056180
## Class: 4_specific 0.5847458 0.8740360 0.7684779 0.7464325
## Class: 6_holistic 0.4216216 0.9777035 0.4382022 0.9761799
## Class: 7_idiomatic 0.0000000 1.0000000 NaN 0.9992505
## Class: 8_nobasis 0.0000000 0.9985931 0.0000000 0.9892784
## Precision Recall F1 Prevalence
## Class: 1_pointer 0.0000000 0.0000000 NaN 0.0097430407
## Class: 2_copyedit 0.5983393 0.8415082 0.6993899 0.3208779443
## Class: 3_general 0.6574648 0.6204147 0.6384026 0.2013918630
## Class: 4_specific 0.7684779 0.5847458 0.6641388 0.4169164882
## Class: 6_holistic 0.4382022 0.4216216 0.4297521 0.0396145610
## Class: 7_idiomatic NA 0.0000000 NA 0.0007494647
## Class: 8_nobasis 0.0000000 0.0000000 NaN 0.0107066381
## Detection Rate Detection Prevalence Balanced Accuracy
## Class: 1_pointer 0.00000000 0.001927195 0.4990269
## Class: 2_copyedit 0.27002141 0.451284797 0.7872999
## Class: 3_general 0.12494647 0.190042827 0.7694512
## Class: 4_specific 0.24379015 0.317237687 0.7293909
## Class: 6_holistic 0.01670236 0.038115632 0.6996625
## Class: 7_idiomatic 0.00000000 0.000000000 0.5000000
## Class: 8_nobasis 0.00000000 0.001391863 0.4992965# This block outputs a single line table with overall accuracy, kappa, lower and higher limits of accuracy, accuracy null value, P value for accuracy, and Mcnemar P value for Structure x All Data.
print ("All, Structure")## [1] "All, Structure"conf.mat4$overall## Accuracy Kappa AccuracyLower AccuracyUpper AccuracyNull
## 0.6554604 0.4956341 0.6457220 0.6651018 0.4169165
## AccuracyPValue McnemarPValue
## 0.0000000 NaN# This block outputs JUST the overall accuracy value for Structure x All Data.
print ("All, Structure")## [1] "All, Structure"conf.mat4$overall['Accuracy']## Accuracy
## 0.6554604Output for Review, Analysis
The code block below creates a CSV file containing the original unmodified TA comments, the subject and structure labels assigned by a human observer, and the corresponding predicted categories for each comment based on the Naive Bayes estimator. Results are exported to a CSV file. By default the CSV file should be saved to the current working directory.
#Creates a data frame that compares predicted and actual labels for subject and structure
tallymatrix <- data.frame("text"=c(comments_rawVERB_R[1:9340,]$text),"subject_label"=c(comments_all_labels_subject), "subject_predicted"=c(comments_all_pred_subject),"structure_label"=c(comments_all_labels_structure), "structure_predicted"=c(comments_all_pred_structure))
#Creates a summary counts table that compares hand-coded counts to Bayes-estimated counts.
final.tally <- table("Hand Coded Subject"=tallymatrix$subject_label, "Hand Coded Structure"=tallymatrix$structure_label)
final.tally2 <- table("Bayes Predicted Subject"=tallymatrix$subject_predicted, "Bayes Predicted Structure"=tallymatrix$structure_predicted)
#Write out the final data from the run to a CSV file. For reference, include the randomizer value used in the name of the file.
write.csv(tallymatrix, file = "SandS_Matrix_Randomizer_123.csv")
#Print to console the hand coded versus estimated counts as a 2-way contingency table of subject vs. structure.
final.tally## Hand Coded Structure
## Hand Coded Subject 1 2 3 4 5 6 7
## 1 0 4 107 99 1 0 0
## 2 62 724 776 881 99 3 33
## 3 18 2225 667 2372 66 4 57
## 4 11 44 331 542 204 0 10prop_tally<-prop.table(final.tally)
print(prop_tally)## Hand Coded Structure
## Hand Coded Subject 1 2 3 4
## 1 0.0000000000 0.0004282655 0.0114561028 0.0105995717
## 2 0.0066381156 0.0775160600 0.0830835118 0.0943254818
## 3 0.0019271949 0.2382226981 0.0714132762 0.2539614561
## 4 0.0011777302 0.0047109208 0.0354389722 0.0580299786
## Hand Coded Structure
## Hand Coded Subject 5 6 7
## 1 0.0001070664 0.0000000000 0.0000000000
## 2 0.0105995717 0.0003211991 0.0035331906
## 3 0.0070663812 0.0004282655 0.0061027837
## 4 0.0218415418 0.0000000000 0.0010706638print(line)## [1] "**************************************************************"print(line)## [1] "**************************************************************"final.tally2## Bayes Predicted Structure
## Bayes Predicted Subject 1 2 3 4 5 7
## 1 7 0 90 39 12 4
## 2 0 867 912 781 60 1
## 3 5 3340 552 1602 6 2
## 4 6 8 221 541 278 6prop_tally2<-prop.table(final.tally2)
print(prop_tally2)## Bayes Predicted Structure
## Bayes Predicted Subject 1 2 3
## 1 0.0007494647 0.0000000000 0.0096359743
## 2 0.0000000000 0.0928265525 0.0976445396
## 3 0.0005353319 0.3576017131 0.0591006424
## 4 0.0006423983 0.0008565310 0.0236616702
## Bayes Predicted Structure
## Bayes Predicted Subject 4 5 7
## 1 0.0041755889 0.0012847966 0.0004282655
## 2 0.0836188437 0.0064239829 0.0001070664
## 3 0.1715203426 0.0006423983 0.0002141328
## 4 0.0579229122 0.0297644540 0.0006423983This block is an alternative method to get the same data tables out using tidy format. It seems much less direct, but may be over-written.
#Use broom::tidy to reorganize into a data frame.
tidy_tally1 <-tidy(final.tally)
tidy_tally2 <-tidy(final.tally2)
tidy_tally3 <-tidy(final.tally)
tidy_tally4 <-tidy(final.tally2)
#Use mutate to add a proportion of total column. And yes, it calculates based on subgroups rather than whole column.
prop_tally1<-mutate(tidy_tally1, prop = Freq/sum(Freq))
prop_tally1<-select(prop_tally1, 1,2,4)
prop_tally2<-mutate(tidy_tally2, prop = Freq/sum(Freq))
prop_tally2<-select(prop_tally2, 1,2,4)
prop_tally3<-mutate(tidy_tally3, prop = Freq/sum(Freq))
prop_tally3<-select(prop_tally3, 1,2,4)
prop_tally4<-mutate(tidy_tally4, prop = Freq/sum(Freq))
prop_tally4<-select(prop_tally4, 1,2,4)
#Spread tidy data into a readable table
spread(prop_tally1,key=Hand.Coded.Structure, value=prop)spread(prop_tally2,key=Bayes.Predicted.Structure, value=prop)spread(prop_tally3,key=Hand.Coded.Subject, value=prop)spread(prop_tally4,key=Bayes.Predicted.Subject, value=prop)#Calculate the sum of values for each group.
proptallysum1<-prop_tally1 %>% group_by(Hand.Coded.Subject) %>% summarize(total=sum(prop))
proptallysum1proptallysum2<-prop_tally2 %>% group_by(Bayes.Predicted.Subject) %>% summarize(total=sum(prop))
proptallysum2proptallysum3<-prop_tally3 %>% group_by(Hand.Coded.Structure) %>% summarize(total=sum(prop))
proptallysum3proptallysum4<-prop_tally4 %>% group_by(Bayes.Predicted.Structure) %>% summarize(total=sum(prop))
proptallysum4Comparing Coded and Predicted Categories
These code blocks compare the hand-coded versus NB-predicted frequencies. There are separate comparisons for the Subject and Structure categories, and both tabular and graphical comparisons.
colnames(proptallysum1)[colnames(proptallysum1)=="Hand.Coded.Subject"] <- "Subject"
colnames(proptallysum1)[colnames(proptallysum1)=="total"] <- "Hand"
colnames(proptallysum2)[colnames(proptallysum2)=="Bayes.Predicted.Subject"] <- "Subject"
colnames(proptallysum2)[colnames(proptallysum2)=="total"] <- "Bayes"
propmerge12<-merge.data.frame(proptallysum1,proptallysum2, all=TRUE)
propmerge12blendSubject = ggplot()+
geom_col(data=proptallysum1, aes(`Subject`, `Hand`, group=1), color="blue", fill="cornflowerblue", alpha=0.5, width=0.4) +
geom_col(data=proptallysum2, aes(`Subject`, `Bayes`, group=1), color="brown", fill="indianred4", alpha=0.5, width=0.4,position = position_nudge(x=0.12))+
xlab("Subject Groups")+
ylab("Fraction of All Comments")+
ggtitle("Comment Subject Frequencies: Coded (blue) vs Predicted (red)")
print(blendSubject)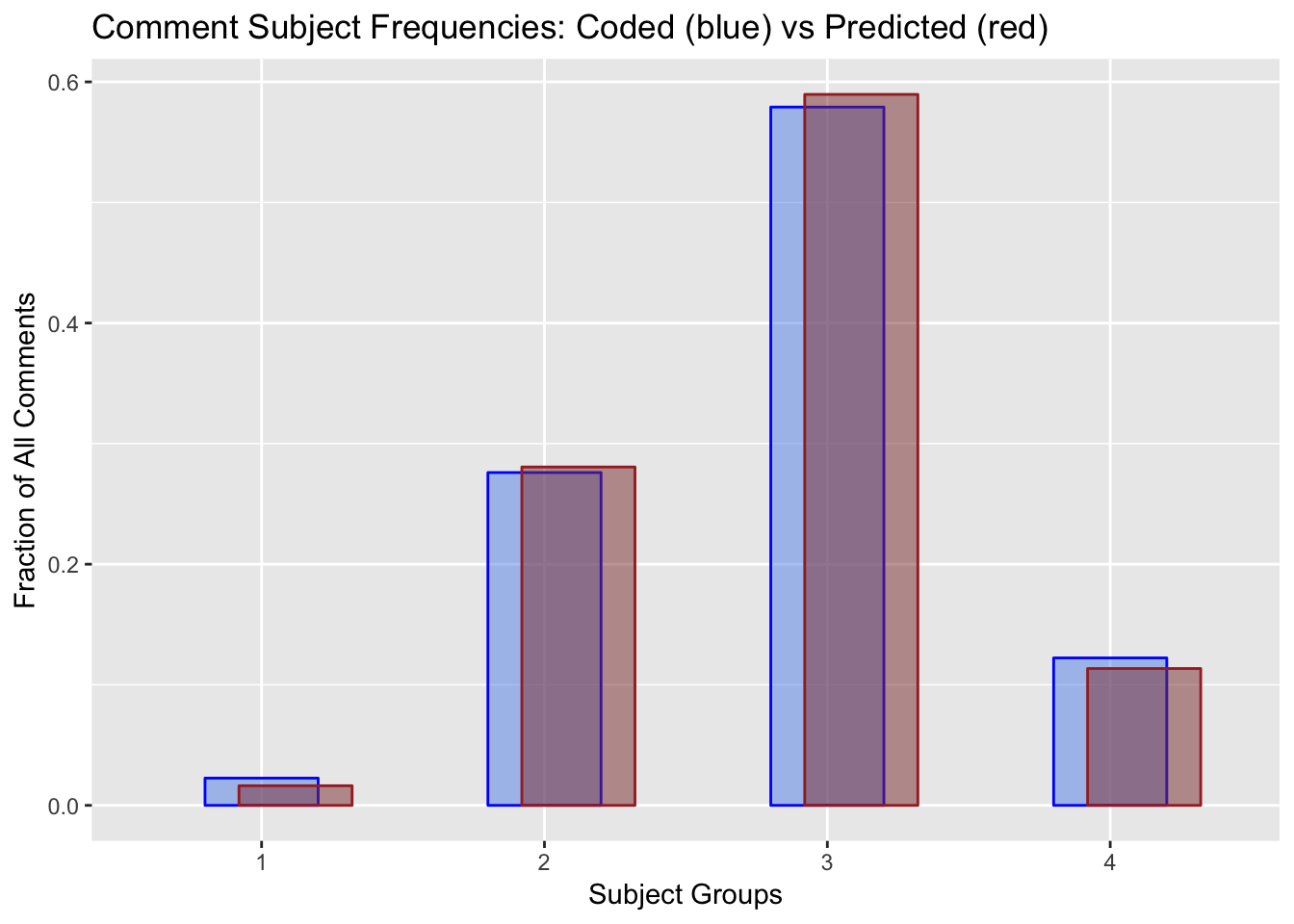
colnames(proptallysum3)[colnames(proptallysum3)=="Hand.Coded.Structure"] <- "Structure"
colnames(proptallysum3)[colnames(proptallysum3)=="total"] <- "Hand"
colnames(proptallysum4)[colnames(proptallysum4)=="Bayes.Predicted.Structure"] <- "Structure"
colnames(proptallysum4)[colnames(proptallysum4)=="total"] <- "Bayes"
propmerge34<-merge.data.frame(proptallysum3,proptallysum4, all=TRUE)
propmerge34blendStructure = ggplot()+
geom_col(data=proptallysum3, aes(`Structure`, `Hand`,group=1), color="springgreen4", fill="springgreen3", alpha=0.5, width=0.4) +
geom_col(data=proptallysum4, aes(`Structure`, `Bayes`, group=1), color="goldenrod3", fill="goldenrod2", alpha=0.5, width=0.4, position = position_nudge(x=0.12))+
xlab("Structure Groups")+
ylab("Fraction of All Comments")+
ggtitle("Comment Structure Frequencies: Coded (green) vs Predicted (orange)")
print(blendStructure)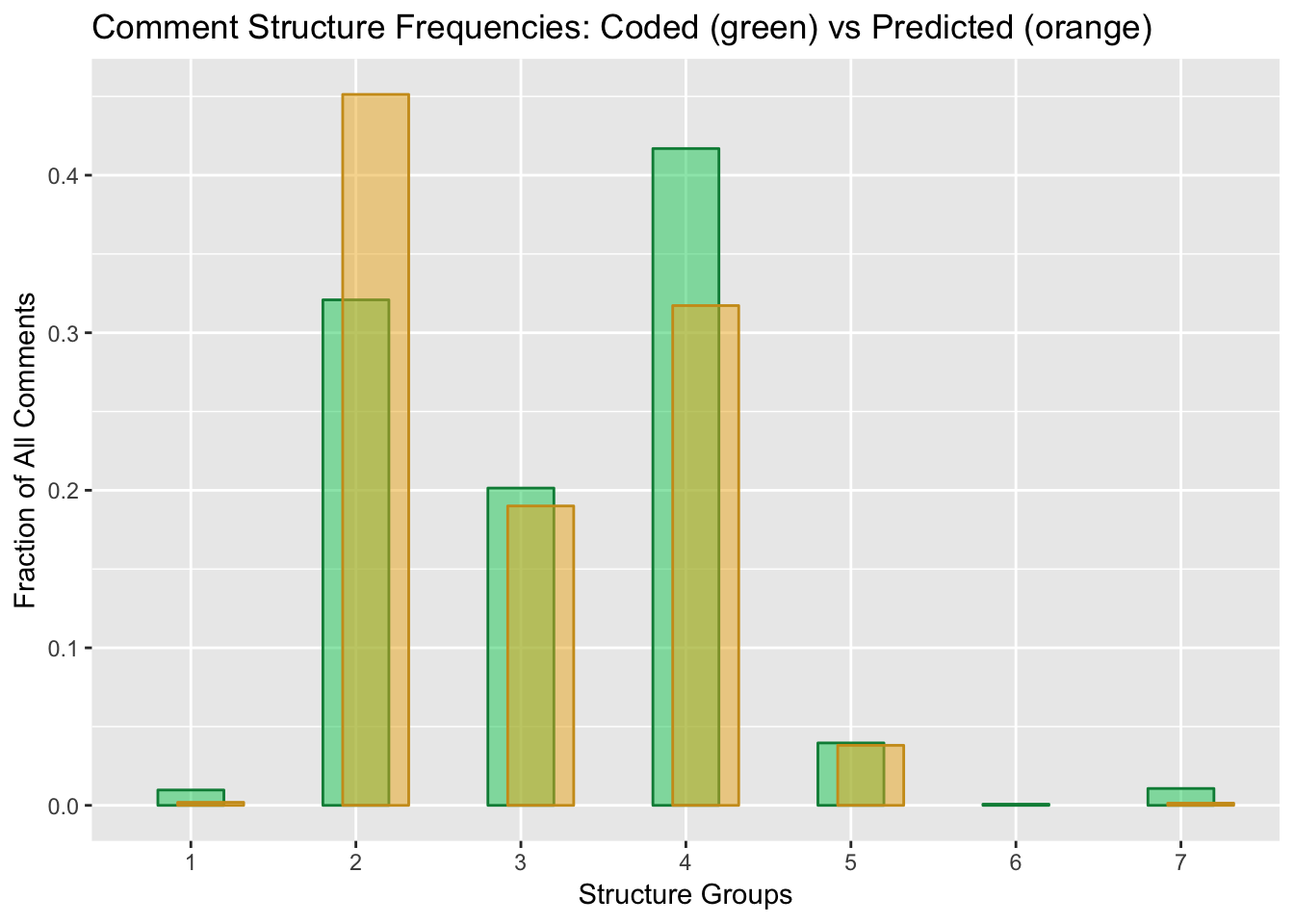
Copyright © 2019 A. Daniel Johnson. All rights reserved.